
Whole Genome de novo Assembly |Supporting de novo assembly training |Course |WGS sequencing | data analysis| Workshop

Selecting Superior De Novo Transcriptome Assemblies: Lessons Learned by Leveraging the Best Plant Genome
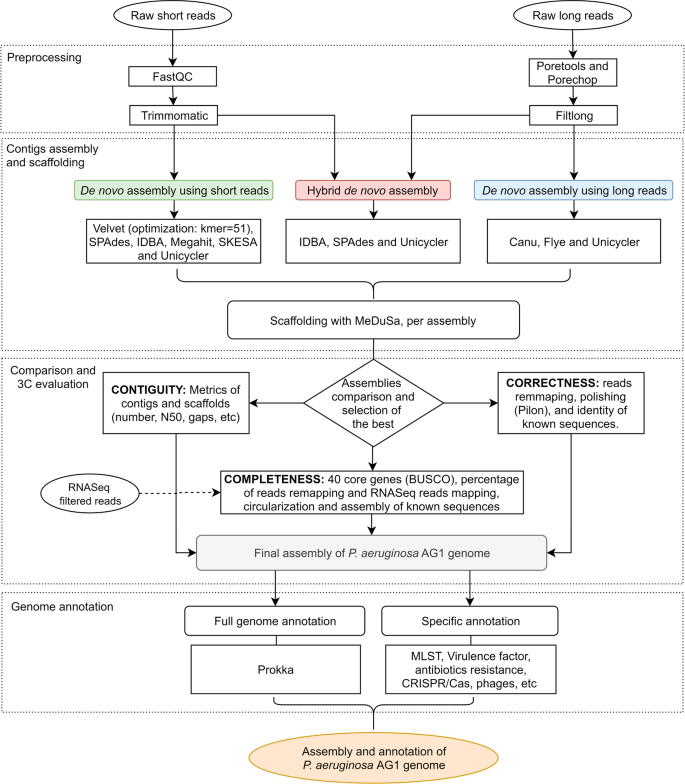
High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers | Scientific Reports
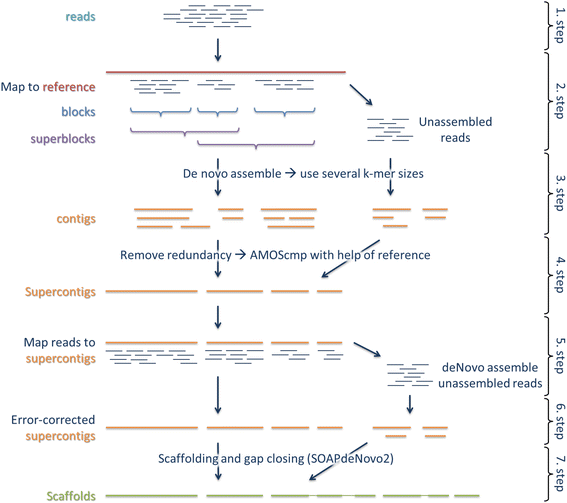
Reference-guided de novo assembly approach improves genome reconstruction for related species | BMC Bioinformatics | Full Text

Rapid hybrid de novo assembly of a microbial genome using only short reads: Corynebacterium pseudotuberculosis I19 as a case study - ScienceDirect

Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions | Nature Communications

Assembly and analysis of a qingke reference genome demonstrate its close genetic relation to modern cultivated barley - Dai - 2018 - Plant Biotechnology Journal - Wiley Online Library
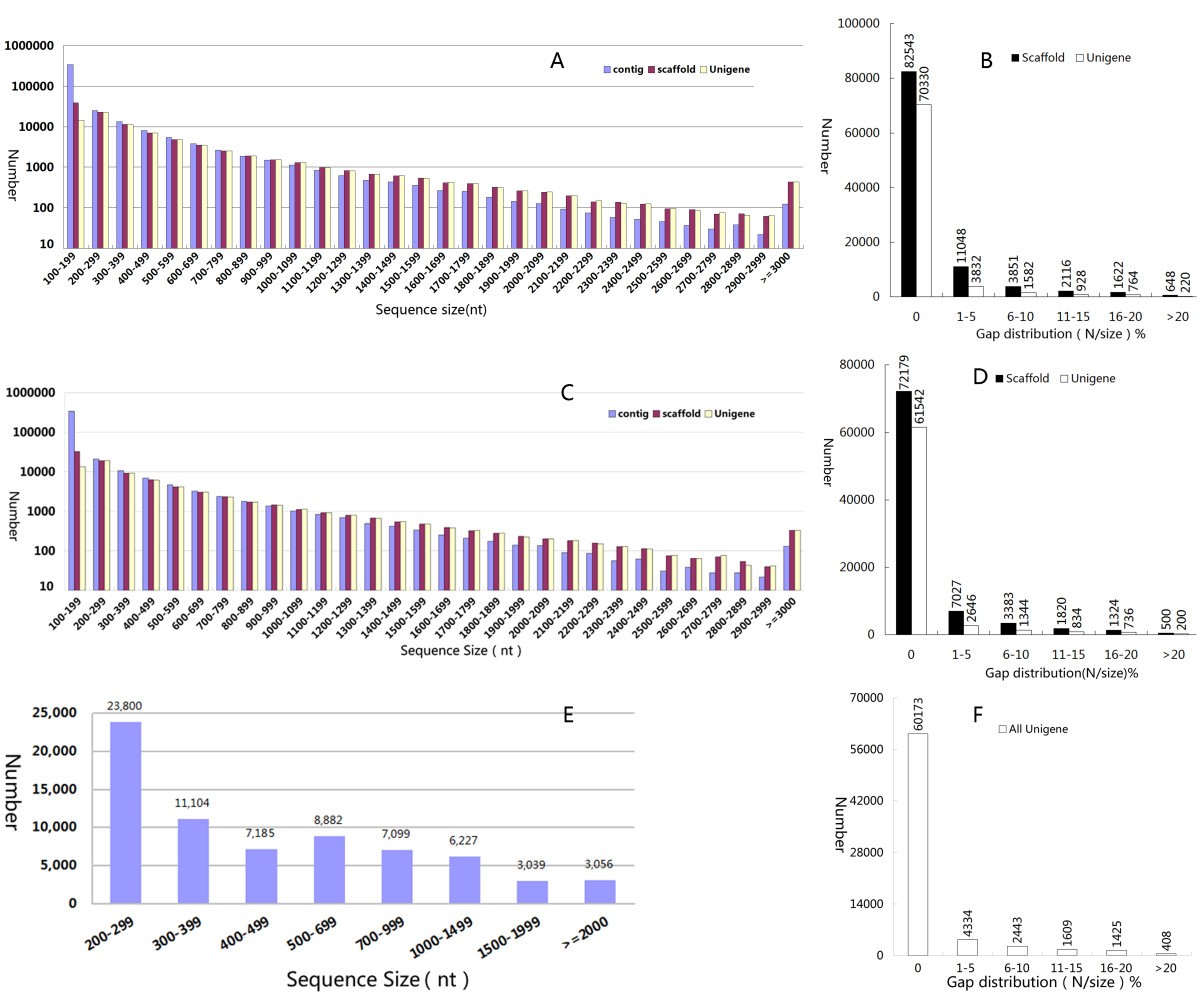
Transcriptome analysis of Sacha Inchi ( Plukenetia volubilis L . ) seeds at two developmental stages | BMC Genomics | Full Text

A comparative analysis of methods for de novo assembly of hymenopteran genomes using either haploid or diploid samples | Scientific Reports

Flow chart of our reference guided genome assembly pipeline. All reads... | Download Scientific Diagram
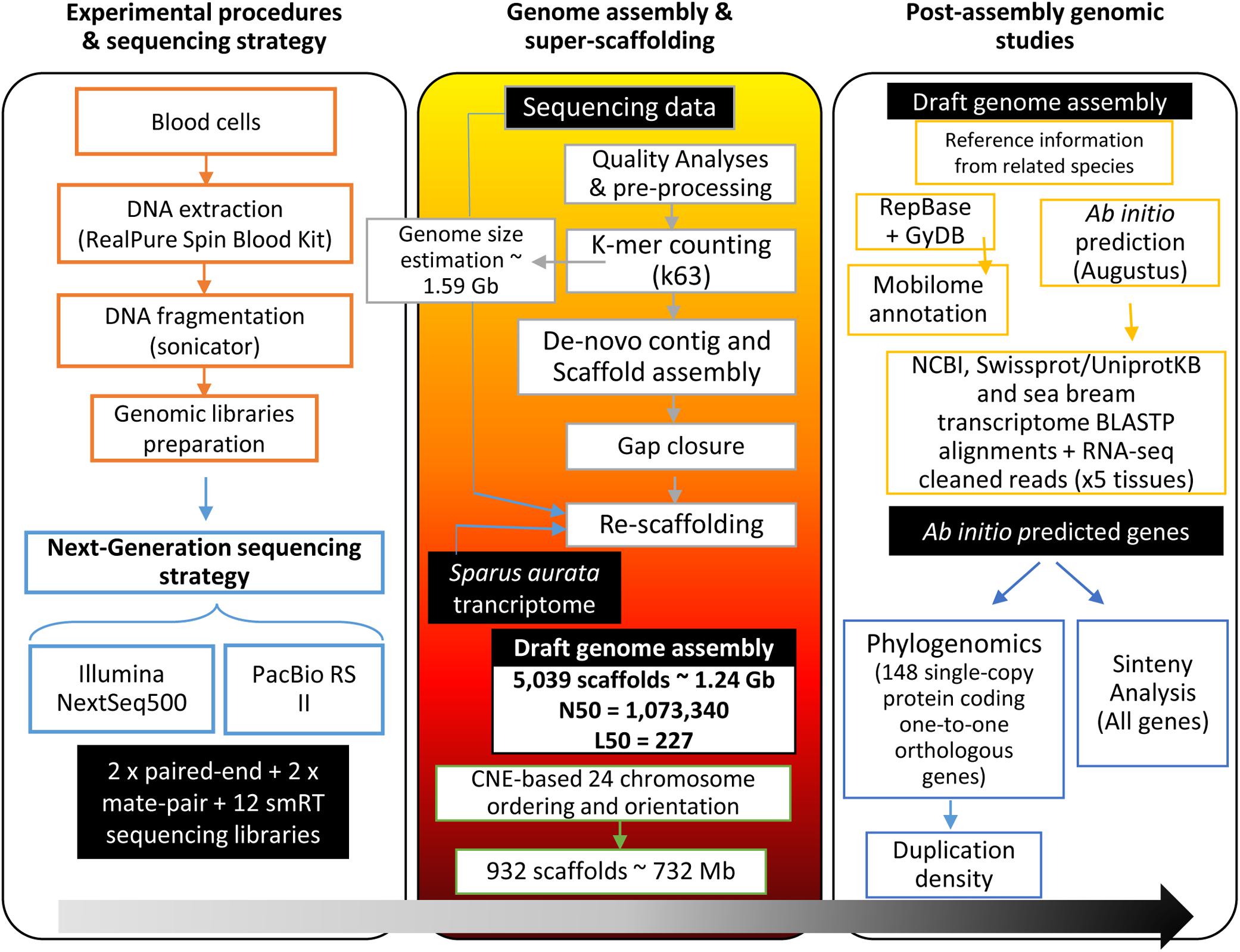
Frontiers | Genome Sequencing and Transcriptome Analysis Reveal Recent Species-Specific Gene Duplications in the Plastic Gilthead Sea Bream (Sparus aurata) | Marine Science
Selecting Superior De Novo Transcriptome Assemblies: Lessons Learned by Leveraging the Best Plant Genome

Closing Human Reference Genome Gaps: Identifying and Characterizing Gap-Closing Sequences | G3: Genes | Genomes | Genetics





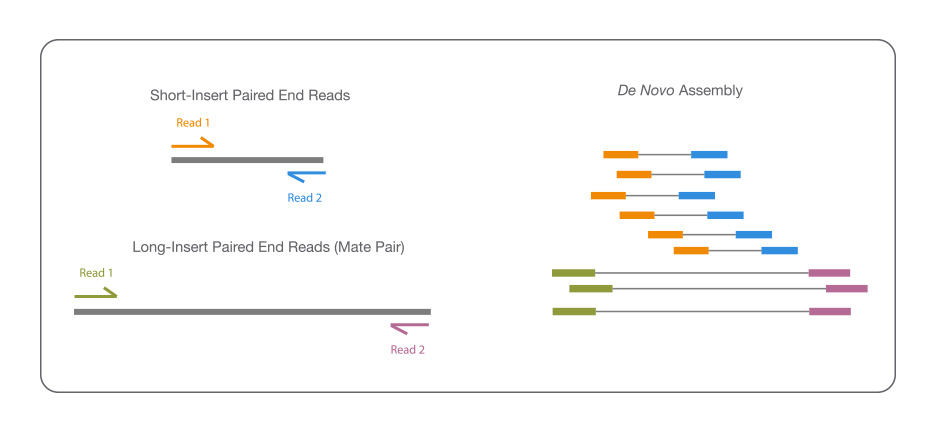


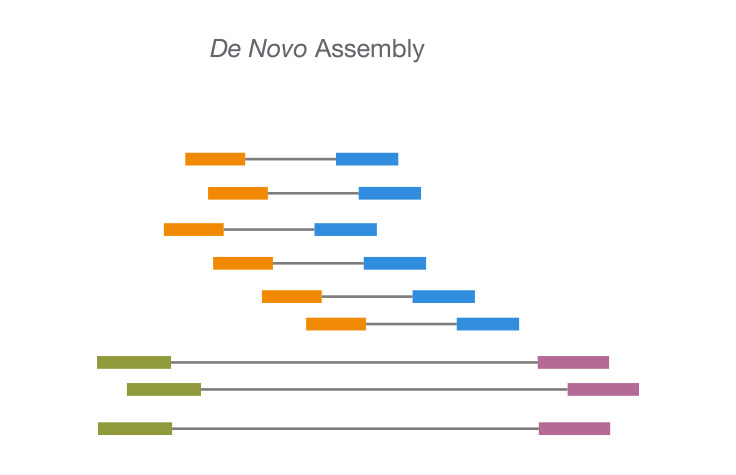
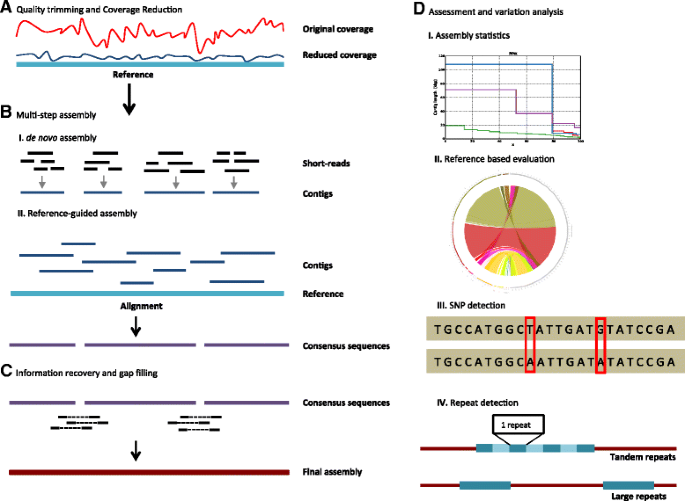

![Optimizing de novo genome assembly from PCR-amplified metagenomes [PeerJ] Optimizing de novo genome assembly from PCR-amplified metagenomes [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2019/6902/1/fig-2-full.png)